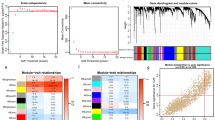
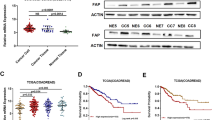
Abstract
Colorectal cancer (CRC) is the third most common malignant disease worldwide, and its incidence is increasing, but the molecular mechanisms of this disease are highly heterogeneous and still far from being fully understood. Increasing evidence suggests that fibrosis mediated by abnormal activation of fibroblasts based in the microenvironment is associated with a poor prognosis. However, the function and pathogenic mechanisms of fibroblasts in CRC remain unclear. Here, combining scrna-seq and clinical specimen data, DAZ Interacting Protein 1 (DZIP1) was found to be expressed on fibroblasts and cancer cells and positively correlated with stromal deposition. Importantly, pseudotime-series analysis showed that DZIP1 levels were up-regulated in malignant transformation of fibroblasts and experimentally confirmed that DZIP1 modulates activation of fibroblasts and promotes epithelial-mesenchymal transition (EMT) in tumor cells. Further studies showed that DZIP1 expressed by tumor cells also has a driving effect on EMT and contributes to the recruitment of more fibroblasts. A similar phenomenon was observed in xenografted nude mice. And it was confirmed in xenograft mice that downregulation of DZIP1 expression significantly delayed tumor formation and reduced tumor size in CRC cells. Taken together, our findings suggested that DZIP1 was a regulator of the CRC mesenchymal phenotype. The revelation of targeting DZIP1 provides a new avenue for CRC therapy.






Similar content being viewed by others
Data Availability
The data and materials in the current study are available from the corresponding author: Wei Huang: huangwei87nj@163.com and Xiao-ping Qian: 20223053@njucm.edu.cn.
References
Biller, L. H., & Schrag, D. (2021). Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA, 325(7), 669–685. https://doi.org/10.1001/jama.2021.0106
Fan, A., Wang, B., Wang, X., Nie, Y., Fan, D., Zhao, X., & Lu, Y. (2021). Immunotherapy in colorectal cancer: Current achievements and future perspective. International Journal of Biological Sciences, 17(14), 3837–3849. https://doi.org/10.7150/ijbs.64077
Li, N., Lu, B., Luo, C., Cai, J., Lu, M., Zhang, Y., Chen, H., & Dai, M. (2021). Incidence, mortality, survival, risk factor and screening of colorectal cancer: A comparison among China, Europe, and northern America. Cancer Letters, 522, 255–268. https://doi.org/10.1016/j.canlet.2021.09.034
Li, J., Ma, X., Chakravarti, D., Shalapour, S., & DePinho, R. A. (2021). Genetic and biological hallmarks of colorectal cancer. Genes & Development, 35(11–12), 787–820. https://doi.org/10.1101/gad.348226.120
Xie, S., Cai, Y., Chen, D., Xiang, Y., Cai, W., Mao, J., & Ye, J. (2022). Single-cell transcriptome analysis reveals heterogeneity and convergence of the tumor microenvironment in colorectal cancer. Frontiers in Immunology, 13, 1003419. https://doi.org/10.3389/fimmu.2022.1003419
Sathe, A., Mason, K., Grimes, S. M., Zhou, Z., Lau, B. T., Bai, X., Su, A., Tan, X., Lee, H., Suarez, C. J., Nguyen, Q., Poultsides, G., Zhang, N. R., & Ji, H. P. (2023). Colorectal cancer metastases in the liver establish immunosuppressive spatial networking between tumor-associated SPP1+ macrophages and fibroblasts. Clinical Cancer Research, 29(1), 244–260. https://doi.org/10.1158/1078-0432.Ccr-22-2041
Wang, H., Tian, T., & Zhang, J. (2021). Tumor-associated macrophages (TAMs) in colorectal cancer (CRC): From mechanism to therapy and prognosis. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms22168470
Kamali Zonouzi, S., Pezeshki, P. S., Razi, S., & Rezaei, N. (2022). Cancer-associated fibroblasts in colorectal cancer. Clinical and Translational Oncology, 24(5), 757–769. https://doi.org/10.1007/s12094-021-02734-2
Giguelay, A., Turtoi, E., Khelaf, L., Tosato, G., Dadi, I., Chastel, T., Poul, M. A., Pratlong, M., Nicolescu, S., Severac, D., Adenis, A., Sgarbura, O., Carrère, S., Rouanet, P., Quenet, F., Ychou, M., Pourquier, D., Colombo, P. E., Turtoi, A., & Colinge, J. (2022). The landscape of cancer-associated fibroblasts in colorectal cancer liver metastases. Theranostics, 12(17), 7624–7639. https://doi.org/10.7150/thno.72853
Ren, J., Ding, L., Zhang, D., Shi, G., Xu, Q., Shen, S., Wang, Y., Wang, T., & Hou, Y. (2018). Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics, 8(14), 3932–3948. https://doi.org/10.7150/thno.25541
McAndrews, K. M., Vázquez-Arreguín, K., Kwak, C., Sugimoto, H., Zheng, X., Li, B., Kirtley, M. L., LeBleu, V. S., & Kalluri, R. (2021). αSMA(+) fibroblasts suppress Lgr5(+) cancer stem cells and restrain colorectal cancer progression. Oncogene, 40(26), 4440–4452. https://doi.org/10.1038/s41388-021-01866-7
Zaborowski, A. M., Winter, D. C., & Lynch, L. (2021). The therapeutic and prognostic implications of immunobiology in colorectal cancer: A review. British Journal of Cancer, 125(10), 1341–1349. https://doi.org/10.1038/s41416-021-01475-x
Sorrentino, C., D’Antonio, L., Fieni, C., Ciummo, S. L., & Di Carlo, E. (2021). Colorectal cancer-associated immune exhaustion involves T and B lymphocytes and conventional NK cells and correlates with a shorter overall survival. Frontiers in Immunology, 12, 778329. https://doi.org/10.3389/fimmu.2021.778329
Liu, Y. J., Li, J. P., Zeng, S. H., Han, M., Liu, S. L., & Zou, X. (2021). DZIP1 expression as a prognostic marker in gastric cancer: A bioinformatics-based analysis. Pharmacogenomics and Personalized Medicine, 14, 1151–1168. https://doi.org/10.2147/pgpm.S325701
Schwend, T., Jin, Z., Jiang, K., Mitchell, B. J., Jia, J., & Yang, J. (2013). Stabilization of speckle-type POZ protein (Spop) by Daz interacting protein 1 (Dzip1) is essential for Gli turnover and the proper output of Hedgehog signaling. Journal of Biological Chemistry, 288(45), 32809–32820. https://doi.org/10.1074/jbc.M113.512962
Sekimizu, K., Nishioka, N., Sasaki, H., Takeda, H., Karlstrom, R. O., & Kawakami, A. (2004). The zebrafish iguana locus encodes Dzip1, a novel zinc-finger protein required for proper regulation of Hedgehog signaling. Development, 131(11), 2521–2532. https://doi.org/10.1242/dev.01059
Yin, Y., Liu, Y., Wang, Y., Li, J., Liang, S., Zhang, W., Ma, Z., Liu, S., & Zou, X. (2023). DZIP1 expressed in fibroblasts and tumor cells may affect immunosuppression and metastatic potential in gastric cancer. International Immunopharmacology, 117, 109886. https://doi.org/10.1016/j.intimp.2023.109886
Yan, W., Deng, Y., Zhang, Y., Luo, J., Lu, D., Wan, Q., Mao, L., & Chen, Y. (2019). DZIP1 promotes proliferation, migration, and invasion of oral squamous carcinoma through the GLI1/3 pathway. Translational Oncology, 12(11), 1504–1515. https://doi.org/10.1016/j.tranon.2019.07.005
Han, L., Wang, S., Wei, C., Fang, Y., Huang, S., Yin, T., Xiong, B., & Yang, C. (2021). Tumour microenvironment: A non-negligible driver for epithelial-mesenchymal transition in colorectal cancer. Expert Reviews in Molecular Medicine, 23, e16. https://doi.org/10.1017/erm.2021.13
Taki, M., Abiko, K., Ukita, M., Murakami, R., Yamanoi, K., Yamaguchi, K., Hamanishi, J., Baba, T., Matsumura, N., & Mandai, M. (2021). Tumor immune microenvironment during epithelial-mesenchymal transition. Clinical Cancer Research, 27(17), 4669–4679. https://doi.org/10.1158/1078-0432.Ccr-20-4459
Celià-Terrassa, T., & Jolly, M. K. (2020). Cancer stem cells and epithelial-to-mesenchymal transition in cancer metastasis. Cold Spring Harbor Perspectives in Medicine. https://doi.org/10.1101/cshperspect.a036905
Kojima, Y., Acar, A., Eaton, E. N., Mellody, K. T., Scheel, C., Ben-Porath, I., Onder, T. T., Wang, Z. C., Richardson, A. L., Weinberg, R. A., & Orimo, A. (2010). Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proceedings of the National Academy of Sciences USA, 107(46), 20009–20014. https://doi.org/10.1073/pnas.1013805107
Wang, Y., Lan, W., Xu, M., Song, J., Mao, J., Li, C., Du, X., Jiang, Y., Li, E., Zhang, R., & Wang, Q. (2021). Cancer-associated fibroblast-derived SDF-1 induces epithelial-mesenchymal transition of lung adenocarcinoma via CXCR4/β-catenin/PPARδ signalling. Cell Death & Disease, 12(2), 214. https://doi.org/10.1038/s41419-021-03509-x
Monteran, L., & Erez, N. (2019). The dark side of fibroblasts: Cancer-associated fibroblasts as mediators of immunosuppression in the tumor microenvironment. Frontiers in Immunology, 10, 1835. https://doi.org/10.3389/fimmu.2019.01835
Mao, X., Xu, J., Wang, W., Liang, C., Hua, J., Liu, J., Zhang, B., Meng, Q., Yu, X., & Shi, S. (2021). Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Molecular Cancer, 20(1), 131. https://doi.org/10.1186/s12943-021-01428-1
Hupfer, A., Brichkina, A., Koeniger, A., Keber, C., Denkert, C., Pfefferle, P., Helmprobst, F., Pagenstecher, A., Visekruna, A., & Lauth, M. (2021). Matrix stiffness drives stromal autophagy and promotes formation of a protumorigenic niche. Proceedings of the National Academy of Sciences USA. https://doi.org/10.1073/pnas.2105367118
Lonsdale, J., Thomas, J., Salvatore, M., Phillips, R., Lo, E., Shad, S., Hasz, R., Walters, G., Garcia, F., Young, N., & Foster, B. (2013). The genotype-tissue expression (GTEx) project. Nature Genetics, 45(6), 580–585. https://doi.org/10.1038/ng.2653
Camps, J., Noël, F., Liechti, R., Massenet-Regad, L., Rigade, S., Götz, L., Hoffmann, C., Amblard, E., Saichi, M., Ibrahim, M. M., Pollard, J., Medvedovic, J., Roider, H. G., & Soumelis, V. (2023). Meta-analysis of human cancer single-cell RNA-Seq datasets using the IMMUcan database. Cancer Research, 83(3), 363–373. https://doi.org/10.1158/0008-5472.Can-22-0074
Clough, E., & Barrett, T. (2016). The gene expression omnibus database. Methods in Molecular Biology, 1418, 93–110. https://doi.org/10.1007/978-1-4939-3578-9_5
Guo, A., Wang, W., Shi, H., Wang, J., & Liu, T. (2019). Identification of hub genes and pathways in a rat model of renal ischemia-reperfusion injury using bioinformatics analysis of the gene expression omnibus (GEO) dataset and integration of gene expression profiles. Medical Science Monitor, 25, 8403–8411.
Zhang, L., Li, X., Zhang, J., & Xu, G. (2021). Prognostic implication and oncogenic role of PNPO in pan-cancer. Front Cell Dev Biol, 9, 763674. https://doi.org/10.3389/fcell.2021.763674
Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., Li, B., & Liu, X. S. (2020). TIMER20 for analysis of tumor-infiltrating immune cells. Nucleic Acids Research, 48(W1), W509–W514. https://doi.org/10.1093/nar/gkaa407
Ren, N., Liang, B., & Li, Y. (2020). Identification of prognosis-related genes in the tumor microenvironment of stomach adenocarcinoma by TCGA and GEO datasets. Bioscience Reports. https://doi.org/10.1042/bsr20200980
Tang, S., Liu, Y., & Liu, B. (2022). Integrated bioinformatics analysis reveals marker genes and immune infiltration for pulmonary arterial hypertension. Science and Reports, 12(1), 10154. https://doi.org/10.1038/s41598-022-14307-6
Gaudet, P., Škunca, N., Hu, J. C., & Dessimoz, C. (2017). Primer on the gene ontology. Methods in Molecular Biology, 1446, 25–37. https://doi.org/10.1007/978-1-4939-3743-1_3
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., & Kanehisa, M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 27(1), 29–34. https://doi.org/10.1093/nar/27.1.29
Chen, C., Wang, C., Pang, R., Liu, H., Yin, W., Chen, J., & Tao, L. (2023). Comprehensive single-cell transcriptomic and proteomic analysis reveals NK cell exhaustion and unique tumor cell evolutionary trajectory in non-keratinizing nasopharyngeal carcinoma. Journal of Translational Medicine, 21(1), 278. https://doi.org/10.1186/s12967-023-04112-8
Zhang, Z., Zhu, H., Zhao, C., Liu, D., Luo, J., Ying, Y., & Zhong, Y. (2023). DDIT4 promotes malignancy of head and neck squamous cell carcinoma. Molecular Carcinogenesis, 62(3), 332–347. https://doi.org/10.1002/mc.23489
Hu, J., Jiang, Y., Wei, Q., Li, B., Xu, S., Wei, G., Li, P., Chen, W., Lv, W., Xiao, X., Lu, Y., & Huang, X. (2022). Development of a cancer-associated fibroblast-related prognostic model in breast cancer via bulk and single-cell RNA sequencing. BioMed Research International, 2022, 2955359. https://doi.org/10.1155/2022/2955359
Zhang, Z., Wang, Z. X., Chen, Y. X., Wu, H. X., Yin, L., Zhao, Q., Luo, H. Y., Zeng, Z. L., Qiu, M. Z., & Xu, R. H. (2022). Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response. Genome Medicine, 14(1), 45. https://doi.org/10.1186/s13073-022-01050-w
Long, X., Xiong, W., Zeng, X., Qi, L., Cai, Y., Mo, M., Jiang, H., Zhu, B., Chen, Z., & Li, Y. (2019). Cancer-associated fibroblasts promote cisplatin resistance in bladder cancer cells by increasing IGF-1/ERβ/Bcl-2 signalling. Cell Death & Disease, 10(5), 375. https://doi.org/10.1038/s41419-019-1581-6
Wang, X., Lou, Q., Fan, T., Zhang, Q., Yang, X., Liu, H., & Fan, R. (2023). Copper transporter Ctr1 contributes to enhancement of the sensitivity of cisplatin in esophageal squamous cell carcinoma. Translational Oncology, 29, 101626. https://doi.org/10.1016/j.tranon.2023.101626
Chen, J., Harding, S. M., Natesan, R., Tian, L., Benci, J. L., Li, W., Minn, A. J., Asangani, I. A., & Greenberg, R. A. (2020). Cell cycle checkpoints cooperate to suppress DNA- and RNA-associated molecular pattern recognition and anti-tumor immune responses. Cell Reports, 32(9), 108080. https://doi.org/10.1016/j.celrep.2020.108080
Liu, Y. J., Li, J. P., Zhang, Y., Nie, M. J., Zhang, Y. H., Liu, S. L., & Zou, X. (2021). FSTL3 is a prognostic biomarker in gastric cancer and is correlated with M2 macrophage infiltration. Oncotargets and Therapy, 14, 4099–4117. https://doi.org/10.2147/ott.S314561
Wang, K., Zhang, T., Lei, Y., Li, X., Jiang, J., Lan, J., Liu, Y., Chen, H., Gao, W., Xie, N., Chen, Q., Zhu, X., Liu, X., Xie, K., Peng, Y., Nice, E. C., Wu, M., Huang, C., & Wei, Y. (2018). Identification of ANXA2 (annexin A2) as a specific bleomycin target to induce pulmonary fibrosis by impeding TFEB-mediated autophagic flux. Autophagy, 14(2), 269–282. https://doi.org/10.1080/15548627.2017.1409405
Bozoky, B., Szekely, L., Ernberg, I., & Savchenko, A. (2022). AtlasGrabber: A software facilitating the high throughput analysis of the human protein atlas online database. BMC Bioinformatics, 23(1), 546. https://doi.org/10.1186/s12859-022-05097-9
De Martino, D., & Bravo-Cordero, J. J. (2023). Collagens in cancer: Structural regulators and guardians of cancer progression. Cancer Research, 83(9), 1386–1392. https://doi.org/10.1158/0008-5472.Can-22-2034
Hulikova, A., Black, N., Hsia, L. T., Wilding, J., Bodmer, W. F., & Swietach, P. (2016). Stromal uptake and transmission of acid is a pathway for venting cancer cell-generated acid. Proceedings of the National Academy of Sciences USA, 113(36), E5344-5353. https://doi.org/10.1073/pnas.1610954113
Calvo, F., Ege, N., Grande-Garcia, A., Hooper, S., Jenkins, R. P., Chaudhry, S. I., Harrington, K., Williamson, P., Moeendarbary, E., Charras, G., & Sahai, E. (2013). Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nature Cell Biology, 15(6), 637–646. https://doi.org/10.1038/ncb2756
Doğan, A. (2019). Apelin receptor (Aplnr) signaling promotes fibroblast migration. Tissue and Cell, 56, 98–106. https://doi.org/10.1016/j.tice.2019.01.003
Hosein, A. N., Brekken, R. A., & Maitra, A. (2020). Pancreatic cancer stroma: An update on therapeutic targeting strategies. Nature Reviews Gastroenterology & Hepatology, 17(8), 487–505. https://doi.org/10.1038/s41575-020-0300-1
Liu, Y. J., Han, M., Li, J. P., Zeng, S. H., Ye, Q. W., Yin, Z. H., Liu, S. L., & Zou, X. (2022). An analysis regarding the association between connexins and colorectal cancer (CRC) tumor microenvironment. Journal of Inflammation Research, 15, 2461–2476. https://doi.org/10.2147/jir.S361362
Mao, X. Y., Li, Q. Q., Gao, Y. F., Zhou, H. H., Liu, Z. Q., & Jin, W. L. (2016). Gap junction as an intercellular glue: Emerging roles in cancer EMT and metastasis. Cancer Letters, 381(1), 133–137. https://doi.org/10.1016/j.canlet.2016.07.037
Rudnick, J. A., Monkkonen, T., Mar, F. A., Barnes, J. M., Starobinets, H., Goldsmith, J., Roy, S., Bustamante Eguiguren, S., Weaver, V. M., & Debnath, J. (2021). Autophagy in stromal fibroblasts promotes tumor desmoplasia and mammary tumorigenesis. Genes & Development, 35(13–14), 963–975. https://doi.org/10.1101/gad.345629.120
Walterskirchen, N., Müller, C., Ramos, C., Zeindl, S., Stang, S., Herzog, D., Sachet, M., Schimek, V., Unger, L., Gerakopoulos, V., Hengstschläger, M., Bachleitner-Hofmann, T., Bergmann, M., Dolznig, H., & Oehler, R. (2022). Metastatic colorectal carcinoma-associated fibroblasts have immunosuppressive properties related to increased IGFBP2 expression. Cancer Letters, 540, 215737. https://doi.org/10.1016/j.canlet.2022.215737
Wang, F., Lin, H., Su, Q., & Li, C. (2022). Cuproptosis-related lncRNA predict prognosis and immune response of lung adenocarcinoma. World Journal of Surgical Oncology, 20(1), 275. https://doi.org/10.1186/s12957-022-02727-7
Galon, J., & Bruni, D. (2019). Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nature Reviews Drug Discovery, 18(3), 197–218. https://doi.org/10.1038/s41573-018-0007-y
Asif, P. J., Longobardi, C., Hahne, M., & Medema, J. P. (2021). The role of cancer-associated fibroblasts in cancer invasion and metastasis. Cancers (Basel). https://doi.org/10.3390/cancers13184720
Yoshida, G. J. (2020). Regulation of heterogeneous cancer-associated fibroblasts: The molecular pathology of activated signaling pathways. Journal of Experimental & Clinical Cancer Research, 39(1), 112. https://doi.org/10.1186/s13046-020-01611-0
Arnold, C. R., Lamont, R. E., Walker, J. T., Spice, P. J., Chan, C. K., Ho, C. Y., & Childs, S. J. (2015). Comparative analysis of genes regulated by Dzip1/iguana and hedgehog in zebrafish. Developmental Dynamics, 244(2), 211–223. https://doi.org/10.1002/dvdy.24237
Wang, C., Low, W. C., Liu, A., & Wang, B. (2013). Centrosomal protein DZIP1 regulates Hedgehog signaling by promoting cytoplasmic retention of transcription factor GLI3 and affecting ciliogenesis. Journal of Biological Chemistry, 288(41), 29518–29529. https://doi.org/10.1074/jbc.M113.492066
Li, C., & Geng, C. (2023). GLIS family zinc finger 3 promotes triple-negative breast cancer progression by inducing cell proliferation, migration and invasion, and activating the NF-κB signaling pathway. Biological and Pharmaceutical Bulletin, 46(2), 209–218. https://doi.org/10.1248/bpb.b22-00595
Liu, Z., Liu, L., Qi, Y., Li, H., & Pan, S. (2021). GLIS family zinc finger 3 promoting cell malignant behaviors and NF-κB signaling in glioma. Brain Research, 1770, 147623. https://doi.org/10.1016/j.brainres.2021.147623
Cheng, B., Yu, Q., & Wang, W. (2023). Intimate communications within the tumor microenvironment: Stromal factors function as an orchestra. Journal of Biomedical Science, 30(1), 1. https://doi.org/10.1186/s12929-022-00894-z
Liao, Z., Tan, Z. W., Zhu, P., & Tan, N. S. (2019). Cancer-associated fibroblasts in tumor microenvironment—Accomplices in tumor malignancy. Cellular Immunology, 343, 103729. https://doi.org/10.1016/j.cellimm.2017.12.003
Zhang, Y., & Zhang, Z. (2020). The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cellular & Molecular Immunology, 17(8), 807–821. https://doi.org/10.1038/s41423-020-0488-6
Chen, Y., McAndrews, K. M., & Kalluri, R. (2021). Clinical and therapeutic relevance of cancer-associated fibroblasts. Nature Reviews Clinical Oncology, 18(12), 792–804. https://doi.org/10.1038/s41571-021-00546-5
Davidson, S., Coles, M., Thomas, T., Kollias, G., Ludewig, B., Turley, S., Brenner, M., & Buckley, C. D. (2021). Fibroblasts as immune regulators in infection, inflammation and cancer. Nature Reviews Immunology, 21(11), 704–717. https://doi.org/10.1038/s41577-021-00540-z
Chen, Y. F., Yu, Z. L., Lv, M. Y., Cai, Z. R., Zou, Y. F., Lan, P., Wu, X. J., & Gao, F. (2021). Cancer-associated fibroblasts impact the clinical outcome and treatment response in colorectal cancer via immune system modulation: A comprehensive genome-wide analysis. Molecular Medicine, 27(1), 139. https://doi.org/10.1186/s10020-021-00402-3
Linares, J., Sallent-Aragay, A., Badia-Ramentol, J., Recort-Bascuas, A., Méndez, A., Manero-Rupérez, N., Re, D. L., Rivas, E. I., Guiu, M., Zwick, M., Iglesias, M., Martinez-Ciarpaglini, C., Tarazona, N., Varese, M., Hernando-Momblona, X., Cañellas-Socias, A., Orrillo, M., Garrido, M., Saoudi, N., … Calon, A. (2023). Long-term platinum-based drug accumulation in cancer-associated fibroblasts promotes colorectal cancer progression and resistance to therapy. Nature Communications, 14(1), 746. https://doi.org/10.1038/s41467-023-36334-1
Sandberg, T. P., Stuart, M., Oosting, J., Tollenaar, R., Sier, C. F. M., & Mesker, W. E. (2019). Increased expression of cancer-associated fibroblast markers at the invasive front and its association with tumor-stroma ratio in colorectal cancer. BMC Cancer, 19(1), 284. https://doi.org/10.1186/s12885-019-5462-2
Schnidar, H., Eberl, M., Klingler, S., Mangelberger, D., Kasper, M., Hauser-Kronberger, C., Regl, G., Kroismayr, R., Moriggl, R., Sibilia, M., & Aberger, F. (2009). Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Research, 69(4), 1284–1292. https://doi.org/10.1158/0008-5472.Can-08-2331
Sabbah, M., Emami, S., Redeuilh, G., Julien, S., Prévost, G., Zimber, A., Ouelaa, R., Bracke, M., De Wever, O., & Gespach, C. (2008). Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resistance Updates, 11(4–5), 123–151. https://doi.org/10.1016/j.drup.2008.07.001
Kraman, M., Bambrough, P. J., Arnold, J. N., Roberts, E. W., Magiera, L., Jones, J. O., Gopinathan, A., Tuveson, D. A., & Fearon, D. T. (2010). Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science, 330(6005), 827–830. https://doi.org/10.1126/science.1195300
Peng, Z., Ye, M., Ding, H., Feng, Z., & Hu, K. (2022). Spatial transcriptomics atlas reveals the crosstalk between cancer-associated fibroblasts and tumor microenvironment components in colorectal cancer. Journal of Translational Medicine, 20(1), 302. https://doi.org/10.1186/s12967-022-03510-8
Poggi, A., Varesano, S., & Zocchi, M. R. (2018). How to hit mesenchymal stromal cells and make the tumor microenvironment immunostimulant rather than immunosuppressive. Frontiers in Immunology, 9, 262. https://doi.org/10.3389/fimmu.2018.00262
Funding
The present study was supported by Nanjing Medical Key Foundation (Grant No. ZKX21028), Jiangsu Province Chinese medicine science and technology development key projects (Grant No. ZD202227), and Jiangsu Province Natural Science Foundation Program (Grant No. BK20211007).
Author information
Authors and Affiliations
Contributions
YZ and Y-jL drafted the manuscript and performed all experiments, JM selected collected histological samples and reviewed histological samples, Z-xY assistance in the animal experiments and analyzed the data. X-pQ and WH provided the research funding. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interest
The authors declare no conflicts of interest. All authors contributed to data analysis, drafting, or revising of the article; agree on the journal to which the article is being submitted; provided final approval of the version to be published; agree to be accountable for all aspects of the work.
Ethical Approval
This study was conducted according to the ethical standards contained in the Declaration of Helsinki, and in national and international guidelines. The study’s protocol was approved by the ethics committee of the Jiangsu Province Hospital of Chinese Medicine, and informed consent was obtained from clinicians and patients (2021NL-206-01).
Consent to Participate
Written information was provided and written consent was given to all participants prior to collection of specimens for research purposes. All mouse experiments were performed by the Guide for Care and Use of Laboratory Animals and approved by the Animal Ethics Committee of Jiangsu Province Hospital of Chinese Medicine (2022DW-10-01). Before data collection, the first author obtained written consent from all participants concerning participation and subsequent publication of the study results.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Liu, Yj., Mei, J. et al. An Analysis Regarding the Association Between DAZ Interacting Zinc Finger Protein 1 (DZIP1) and Colorectal Cancer (CRC). Mol Biotechnol (2024). https://doi.org/10.1007/s12033-024-01065-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-024-01065-1